Introduction
AmiGO is an HTML based application that allows users to browse, query, and visualize data from any ontology in OBO format. The AmiGO browser developed for the Gene Ontology (GO) has been modified for use on the Plant Ontology site. AmiGO is an open source software available from the go-dev cvs repository at the Gene Ontology SourceForge site.
Browser Compatibility
AmiGO is compatible with most browser/operating systems that support Cascading Style Sheets (CSS) including:
- FireFox/Mozilla/Netscape 6 and up on Linux, Windows and Mac OS X.
- Internet Explorer on windows but NOT Mac OS X. IE on Mac OS X does not support CSS standards.
- Safari on Mac OS X.
- Konquerer on Linux.
- Opera on Linux, Windows and Mac OS X.
The Main Window
From the Plant Ontology (PO) browser main page you have options to do the following:
- Browse the ontology and view terms
- Search for the ontology and/or associated annotations
- Filter the annotations which are displayed in the browser.
- Perform an advanced search
- Get data from a related website/database
- Request a PO term
Browsing the ontology.
Figure 1 
Plant Ontology (PO) browser
PO browser hompage (Figure 1) is accessed by clicking "Search/Browse the Plant Ontology" at the menu bar of the POC homepage. There is also a search box on the homepage. Click "Browse" in Figure 1 to view the ontology tree structure (Figure 1A).
Figure 1A 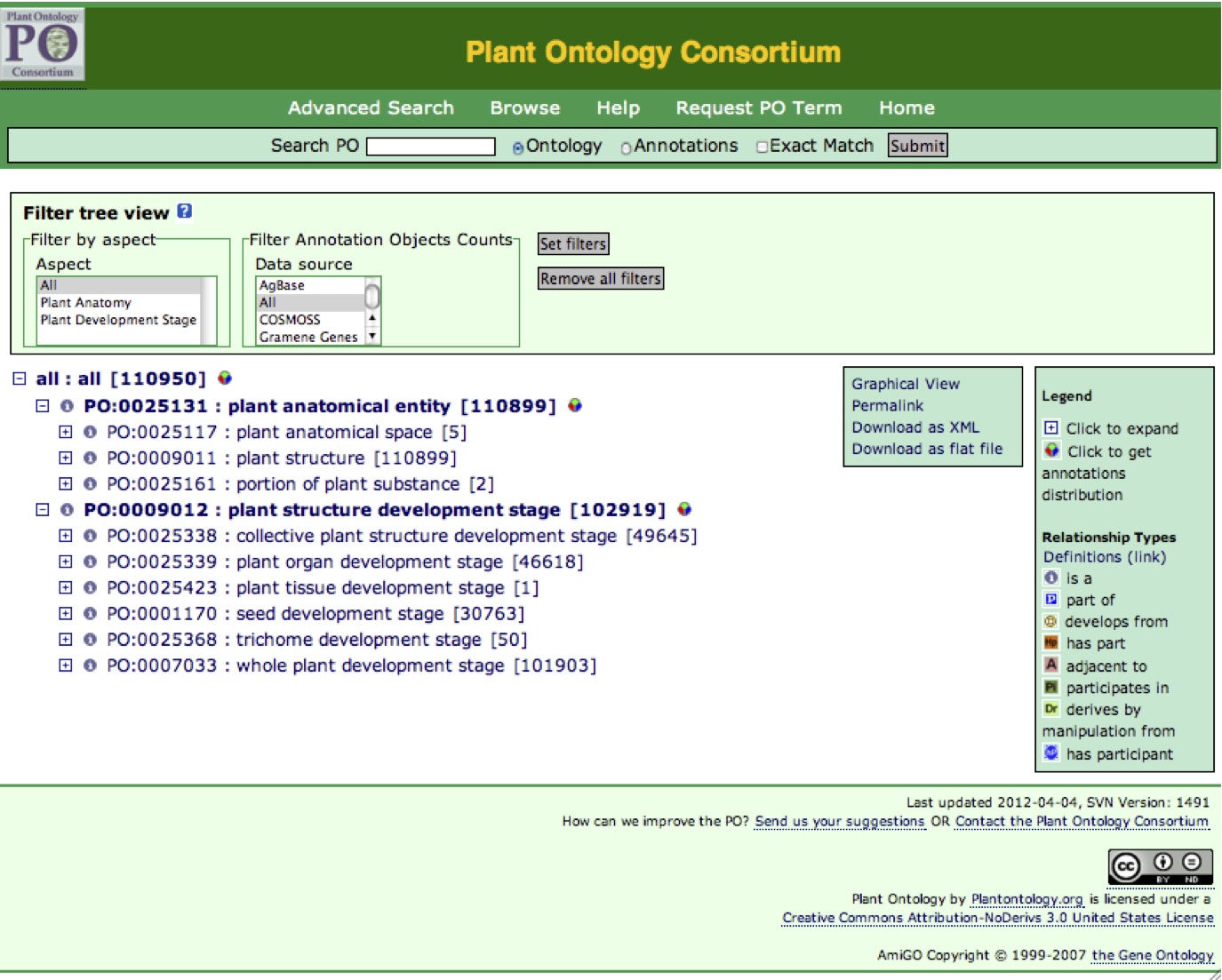
Ontology Tree Structure
The structure of the ontology is presented as a tree of terms (Figure 1A). The top term or node is the highest level of the ontology. Each term is identified by its ontology ID, which has a PO (Plant Ontology) prefix indicating the ontology domain and a seven digit number (e.g., PO:0025131). Each term in the ontology has a unique identifier.
The level of a term in the ontology is represented by its indentation within the tree. In the Plant Ontology, plant anatomical entity (PO:0025131) is at the same level as plant structure development stage (PO:0009012), because these terms are siblings. Furthermore, the relationship of a term to its parent is represented by the icon immediately preceding the term ID. Relationship types include "part of", represented by the blue P icon; "is a", represented by a grey I icon and "develops from", represented by a brown D icon. In the plant ontology, plant anatomical space, plant structure, and portion of plant substance have 'is_a' relationships to the parent term plant anatomical entity.
Filter tree view
The ontology tree can be filtered to limit the display to one aspect or the other (plant anatomical entity or plant structure development stage); the default is to display all terms.
The number of annotations displayed next to each term can be filtered to show the annotations objects from the selected data sources. Click "set filters" to set your display options. If no options are selected, or you click "Remove all filters", the PO browser will display all terms with the full annotation counts.
Understanding term lines
Figure 2 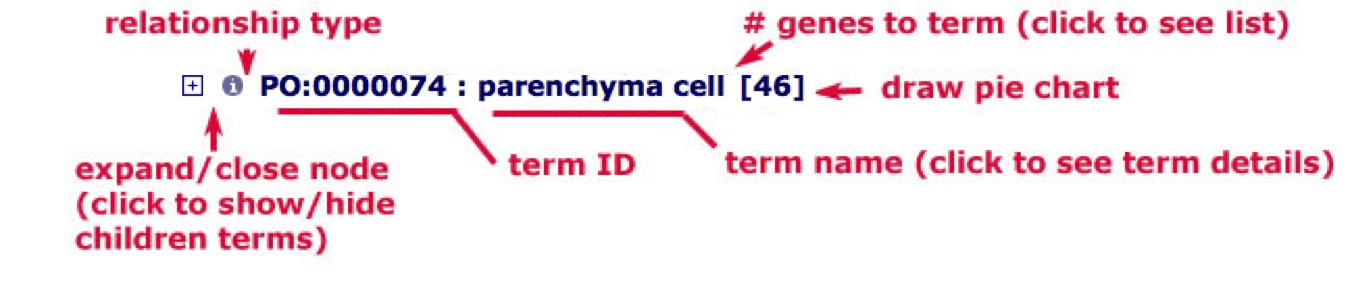
Each line in the tree contains one term, several pieces of information about the term, and some associated functions.
Expanding/Closing term nodes: The term expand/close icon has three forms: a plus sign (+), a minus sign (-), and a period (.). The plus (+) sign indicates that a term has children that are not shown in the current view. The minus (-) sign indicates all of the children of the selected term are shown in the view. The period (.) indicates that the selected term has no children. You can use the expand/close icon to expand a node to see all the children or collapse the node to just the parent term.
Term ID and Term name: The next two items are the term ID and the term name, separated by a colon. To see the full definition of a term, click on the term name to go to the detailed term view.
The status of a term is indicated by font style. If a term is selected in the tree, the name will be shown in bold font.
Number of annotations: Next to the term name is a number in parenthesis. This number represents the total number of annotations to that term. This number can change depending on the search filters applied. When you click the number, a full list of annotation to this term and its children will be displayed, along with brief information on the annotations (Figure 2A).
The PO term is shown in large font at the top of the page. Below the term, links are provided (by clicking the blue arrows) for direct access to the three subsections of the term details page: Term information, Term lineage, and External reference. The type of annotations shown can also be specified using the 'View annotations' filter, by checking one of the options. 'All annotations' will display gene products or other objects annotated to the PO term or to any of its children; 'Direct annotations' will display only gene products or other objects annotated directly to the term.
Figure 2A 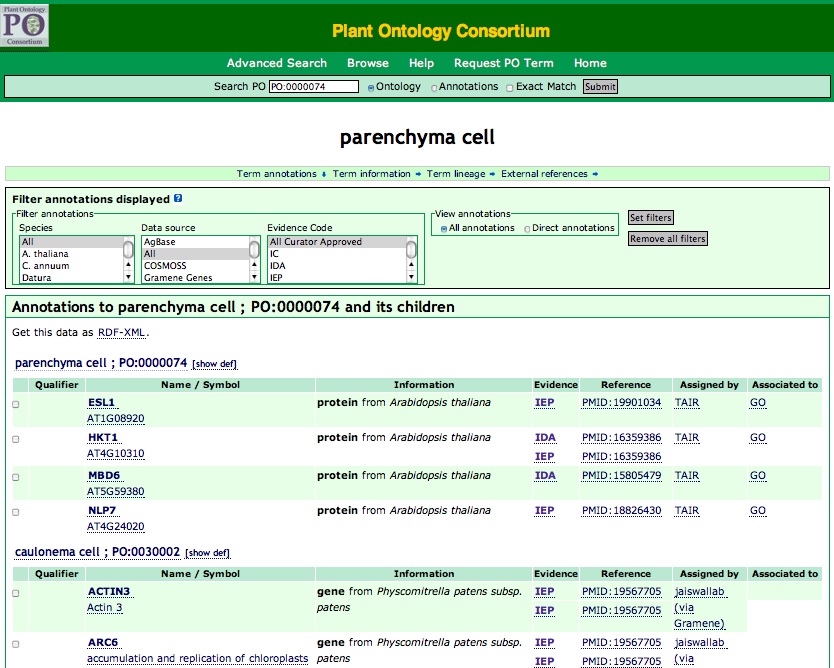
Search the ontology for terms or associated annotations (e.g., gene products).
The ontology can be queried using the search options available in the menu bar at the top of all pages in the PO browser.
Figure 3 ![]()
The database query works by entering a query string into the text input box and selecting the type of search to perform (exact/inexact) and data type to search for. The default option is a search for ontology terms, meaning a search for the string 'leaf' will retrieve all ontology terms containing the word 'leaf'.
Querying for ontology terms
The text string used for queries treats each word in the string as a complete phrase. A search for the string "juvenile leaf" will find terms containing the string "juvenile leaf" rather than "juvenile" OR "leaf". For the broadest search, use a single word. In addition to searching primary term names, the term search also searches alternative names, which are stored in the database as synonyms. For example, a search for the word 'panicle', which is the name of the inflorescence in rice, will return the term ,inflorescence, because panicle is a synonym for this term.
- Enter the term you are searching for into the text input box.
- Select "Ontology" as the data type to search for. This is the default option.
- If you know the exact term you are looking for, choose the "Exact Match" option.
- Click "Submit Query".
The results that match your query are shown in a list (Figure 4). If no results are found, check your spelling or try using simpler terms (leaf vs. juvenile leaf). If you do not find a term in the PO, you can suggest a new term by clicking on the "Request PO term" link in the menu bar at the top of the browser window.
Term search filters
Term searches can be filtered by aspect, to limit the results to a selected branch of the PO (plant anatomical entity or plant structure development stage). The default is to search both aspects. Select more than one aspect to display by control-clicking (command-click on a Mac) the aspect names and click "Set filters". If no options are selected or you click "Reset filters", the PO browser will display results from all aspects.
Figure 4 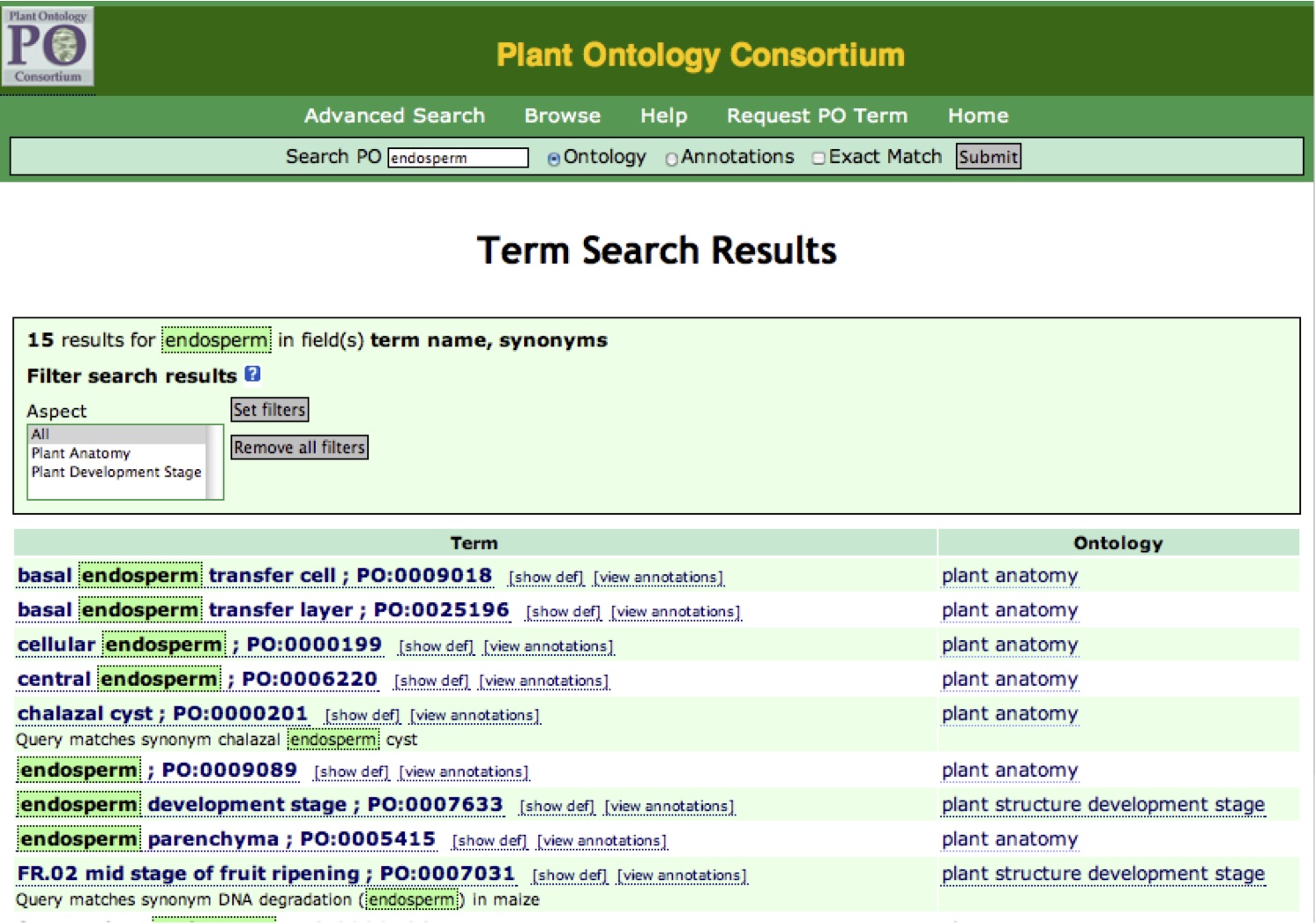
Click on the name of a term to display the term detail view (Figure 9). To the right of the term are "[show def]" and "[view annotations]". Click "[show def]" and the definition of the selected term will appear under the term. Click "[view annotations]" and a list of annotations to the selected term and its childrem will be shown (Figure 4A).
The PO term is shown in large font at the top of the page. Below the term, links are provided (by clicking the blue arrows) for direct access to the three subsections of the term details page: Term information, Term lineage, and External reference.
The type of annotations shown can also be specified. "All annotations" will display gene products annotated to the PO term or to any of its children; "direct annotations" will display only gene products annotated directly to the term.
For genes or gene products which have GO annotations in the GO database, a external link "GO" will appear at the last column of individual annotation, clicking "GO" will open the corresponding GO gene product details page (for example, GO associations to Arabidopsis ABI4 gene)
Figure 4A 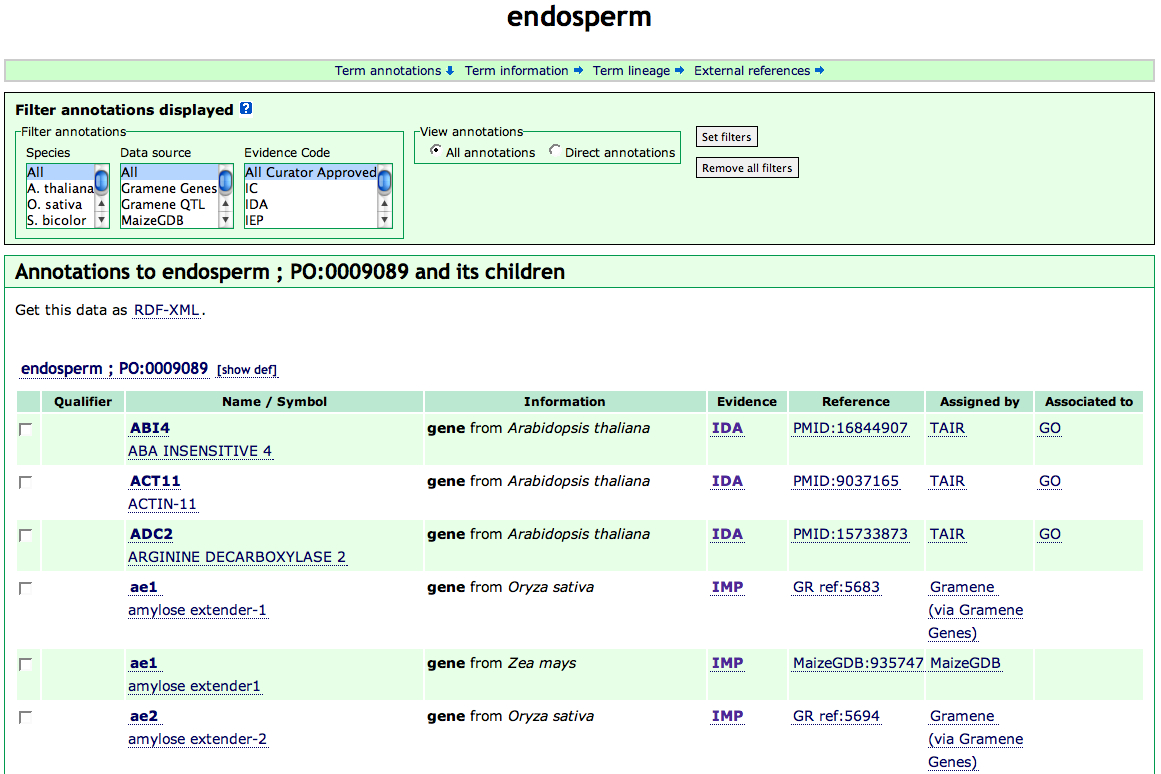
Querying for Annotations
The Plant Ontology Browser allows you to search for "Annotations" (e.g., gene products or other object types) associated to ontology terms in the database (Figure 5). The PO Browser will search annotation object names, symbols, and synonyms, and will return any annotations that contain the query word or phrase.
Figure 5 ![]()
- Enter the name (symbolic or full name ) of the annotation object such as "endosperm".
- Choose the "Annotations" option.
- If you know the exact name or symbol of the annotation object, select an 'Exact Match' checkbox, otherwise the search will be a 'contains' search.
- Click "Submit Query".
The query for annotation object names containing the word "endosperm" is shown in Figure 6.
Figure 6 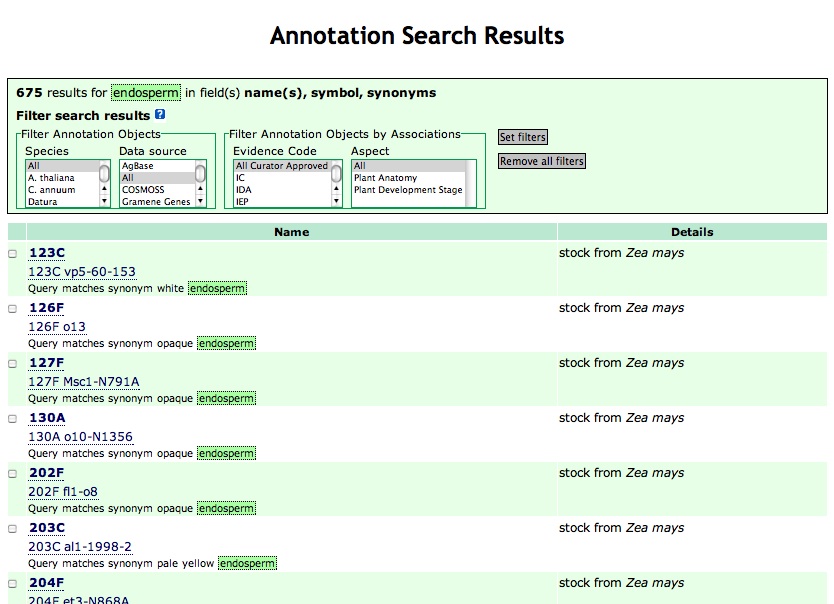
The Query Summary, shown in the top box, lists the search parameters, data filtering options selected, and number of results returned. The main results section lists all of the annotations that matched your query and the species that the annotations are derived from. Clicking on a annotation object name brings you to the annotation object details page (Figure 10).
Annotations Overview and filters
To the left of each row of the table is a check box that can be used for selecting multiple names on the list and to get an "annotations overview" to a set of annotations (Figure 6A). The result can be filtered by "evidence code" and "aspect", to limit the terms shown to those selected filters.
Figure 6A 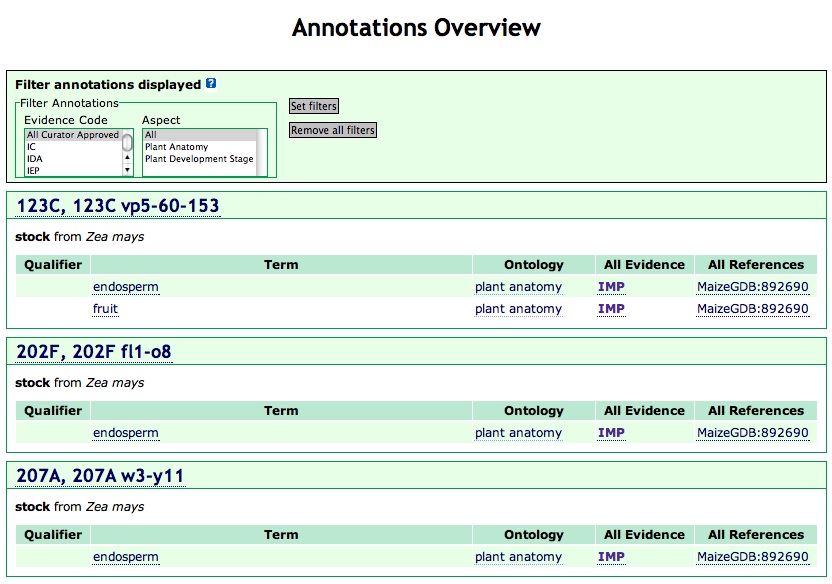
Filtering annotations that are displayed in the browser
The Plant Ontology Browser has four filters that you can use to filter the list of annotations displayed in the browser or included in search results (Figure 7). The default option does not use any filters for retrieving associated data. The filters can be set when performing an advanced database search, or when viewing the ontology tree or term details. One or more filters can be set; the corresponding results will meet the criteria set by ALL the filters in combination.
Using the species filter
The species filter allows you to restrict the display of annotated data to one or more species. This feature is useful if you want to find annotations to a specific organism (say, Zea mays) or a set of organisms. For example, say you wanted to take advantage of maize-rice synteny for identifying candidate genes in maize. Given a phenotype in maize, are there mutants in rice with similar phenotypes?
- Select one or more species to filter. To select multiple species, hold down the CTRL key (PC) or COMMAND key (Mac) while selecting with the mouse.
- Click 'Set Filters'.
Figure 7 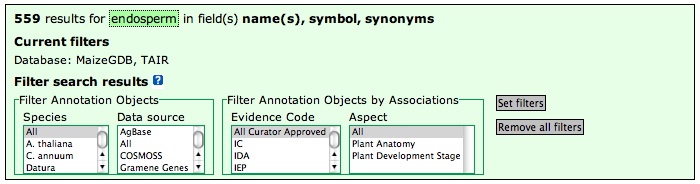
Using the data source filter
This filter allows you to restrict the display of annotated data by contributing database. It functions similarly to the species filter, by limiting the associated data types in the ontology and term detail views, as well as in searches.
Using the evidence code filter
The evidence code filter is a very useful option that allows you to restrict the display of associated data based on the quality of evidence supporting the annotation. The default option is set to 'All Curator Approved'. Curator approved means the association of a term to a data object is based on evidence that was evaluated by a trained biologist. This default option excludes annotations based on electronic unsupervised methods.
Using the aspect filter
You can also filter the annotation objects by "aspect", limiting the anntations shown to those that are annotated to terms in the selected branch of the ontology.
Performing an advanced search
The "Advanced Search" features are accessible by clicking on the 'Advanced Search' link in the PO browser menu bar. These options allow you to query the Plant Ontology with a little more flexibility than a standard search.
Figure 8 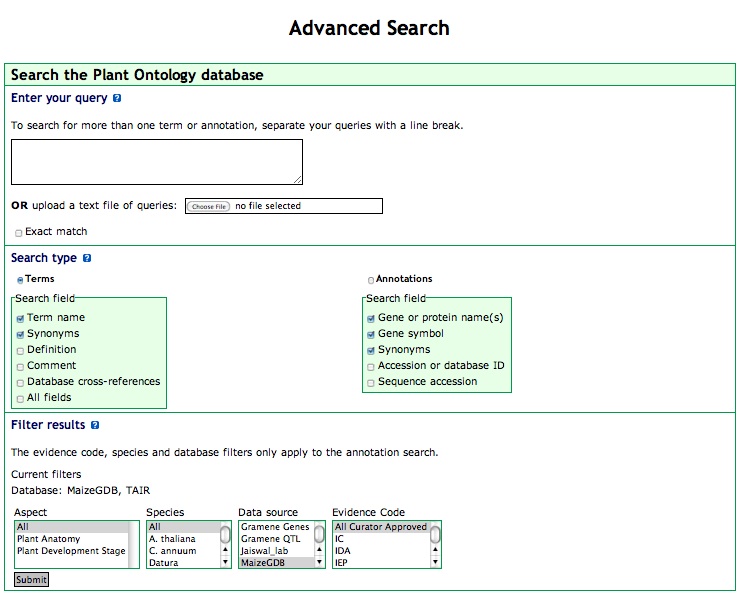
Entering your query: In the text input box, you can enter a single word, as with the Basic Query. You can also enter multiple terms separated by new lines (carriage returns), and the PO browser will search for hits to each of these terms. Alternatively, you can upload a file of terms, where each term is on a single line. The search terms must all be of the same type (PO term names or annotations). Choose 'Terms' or 'Annotations' to search one or the other of these fields. You can also enter phrases into the text box. The entire phrase will be used to search the Plant Ontology.
- Enter the term(s) in the text input box or upload a file of query terms.
- To search for an exact word or phrase, select the "exact match" checkbox.
- Choose the search type. The choices are "Terms" or "Annotations". To specify which data fields the PO browser should search, select one or more of the options in the "search field" section.
- Set the filtering options for annotation search; multiple options can be selected in all the filters; select more than one option by control-clicking (command-click on a Mac) the names.
- Click "Submit Query"
When searching for terms, you can search by:
- Term Name and Synonym.
- Term Name and Definition.
- Ontology ID: e.g. PO:0009074
- All of the above fields
When searching for annotations, the options are:
- Annotation objects Full Name and Symbolic Name
- Full Gene Name
- Gene Symbol
- Synonyms (Gene Name aliases)
Detail Windows
There is a detail pages for every ontology term and annotation object in the PO database. These pages open when you click on the name of a term or an annotation and allow you to explore terms and annotations in depth.
Ontology term details
Overview of "Term Details"
The term detail page displays information associated with a PO term. The PO term is shown in large font at the top of the term details page (Figure 9). Below the term name, links are provided (by clicking the blue arrows) for direct access to the three subsections of the term details page: Term information, Term lineage, and External reference. "Term annotations" is a hyperlink to a page listing all the annotations that are associated with the PO term (Figure 2A, 4A).
Figure 9 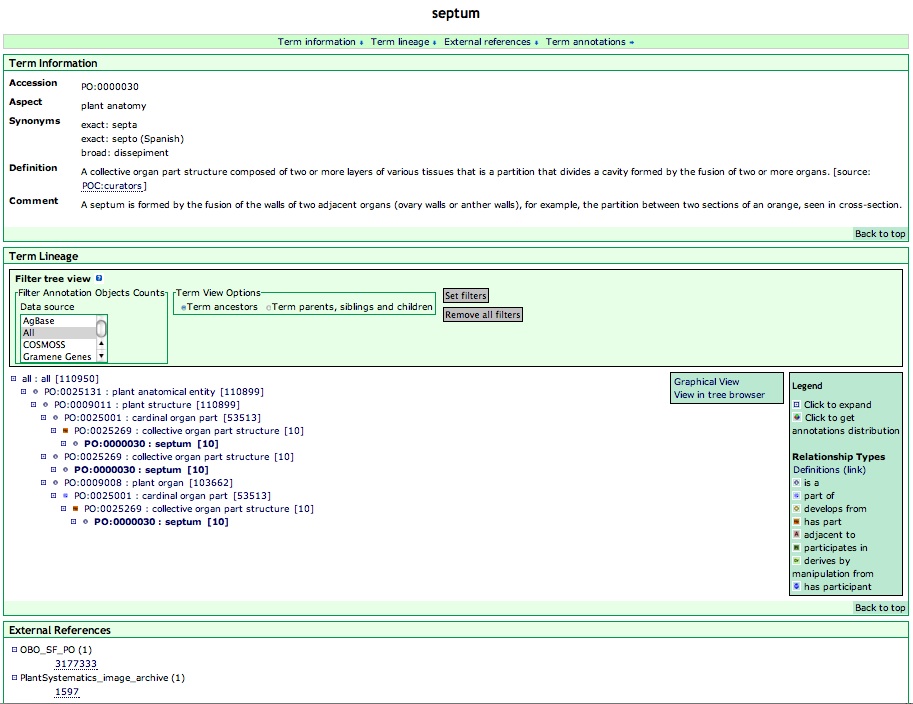
Term Information
"Term Information" displays a list of data fields associated with the PO term: "Accession", "Ontology", "Synonyms", "Definition" and "Comment".
The accession is a unique identifier for the PO term in the PO database. This is always in the format PO:xxxxxxx (a seven digit number).
Synonyms are alternative names for the term. In the PO, synonyms may include more specific instances of a term, for example, a cyme is a specific type of inflorescence and is included as a narrow synonym for the term inflorescence.
The definition is an explanation of what entities should be included in the class for this term. The origin of the definition is indicated in [ ] after "source:".
A wide variety of additional information related to the term is added in the "comment" section, this may include noteworthy biological information related to the term, suggestions for database curators to help them choose the most appropriate term, or historical information on the term.
Term Lineage
The next section on the detail page is "Term Lineage". This section shows the tree view of the term, i.e., the term in relation to other terms in the ontology. The term can be viewed in the context of either 'Term ancestors' or 'Term parents, siblings and children' by checking the preferred checkbox then clicking 'Set filters'. The view will change accordingly. Viewing the Term ancestor is the default view.
The term lineage can also be viewed as a graph, by selecting the option Graphical View from the green box to the right of the tree view.
The number in parenthesis represents the total number of annotations to that term. Click the number, and a full list of annotations to this term and its children will be displayed, along with brief information on the annotation (Figure 4A).
The number of annotations displayed next to each term can be filtered to show the annotations objects from the selected data sources. Click "set filters" to set your display options; if no options are selected or you click "Remove all filters", the PO browser will display all ontologies with full annotation counts.
The tree browser links back to the main term browser (Figure 1A) for further browsing.
External References
This section show cross-references between a PO term and external references. The number of external database entries mapping to the PO term is shown in parentheses after the database abbreviation. This is a single entry in most cases. A ist of identifiers for each database can be viewed by clicking on the (+) icon. Direct links to the database entries are provided when available.
The most common external references are to the PO's SourceForge tracker (OBO_SF_PO), which provides a history of changes to a term, and to Plantsystematics.org (PlantSystematics_image_archive), which provides images of plant structures or development stages.
Annotation Object Details
The Annotation Detail pages are linked from the Annotations Search Results page (Figure 6) and Term Annotations page (Figure 4A). Each contributing database (datasource) submits information about the symbolic name, full name, and aliases for each annotation object. A typical annotation object page is shown in Figure 10.
Figure 10 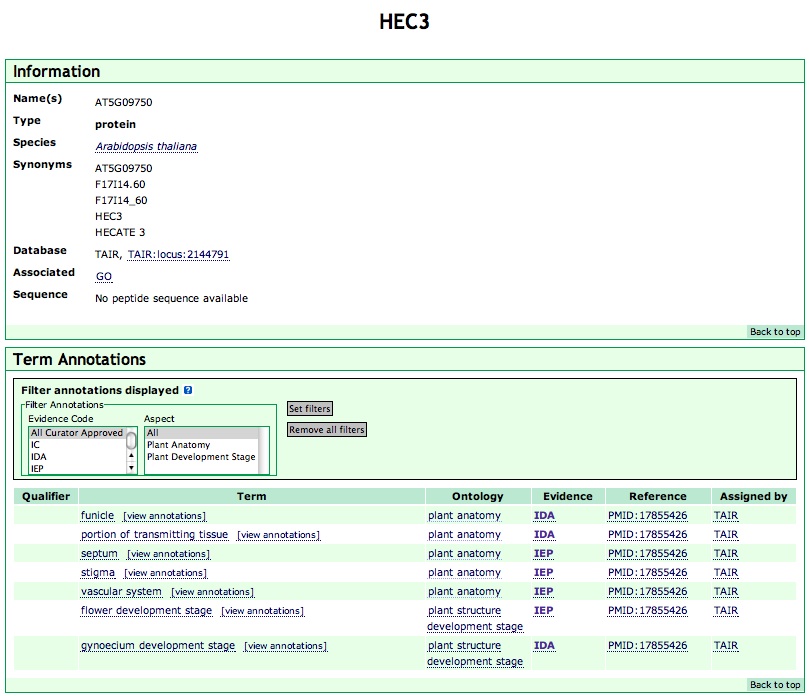
Annotation Details Information
The information section displays the following data associated with the annotation object:
Term Annotations
Any annotation object can be associated to one or more terms in the PO database. A table of the PO terms that the selected gene or gene products has been associated to is displayed here.
The "Qualifier" column is used for flags that modify the interpretation of an annotation. Allowable values are " not", "contributes_to", and "colocalizes_with". More information is in the GO annotation guide.
Clicking the "Term" name will display the "term details page" (Figure 9). Clicking "[view annotations]" in this column will display the "term annotations page" (Figure 4A), with all the gene or gene products annotated to the term.
"Ontology" denotes the aspect that the term belongs to. Clicking on the aspect name will display the PO documentation on that aspect (e.g., plant anatomy).
The "Evidence" can be used to assay the strength of the assertion. Mouse over the evidence code to see the full name of an evidence code, Clicking the selected evidence code will display the evidence code documentation page.
Clicking the "Reference" hyperlink will display the reference used to make the association.
"Assigned by" shows the database or group that made the annotations. If this is different from the database from which the annotation originated, it will state that data were submitted "via" another data source. Clicking on the database link in the "Assigned by" column will open a separate window for that database's page for annotation object.
Term annotations filter
Term annotations can be filtered by "evidence code" and "aspect". Multiple options can be selected in all filters.
Using the links out to contributing databases
There are several places where you can find "Data Source" links to related databases or websites. These links can be used to find additional information about a gene or term.
Data Source links
The "term detail pages" (Figure 9) and "annotation detail pages" (Figure 10) include hyperlinks to data detail pages from the contributing database. The data source listed in this field (e.g. TAIR, Gramene, or MaizeGDB) is a hyperlink. Click on this link to view the database's detail page for the gene, phenotype, allele, or other data type. For each database, the detail pages will provide as much information as they have available about the data type, such as sequences, map locations, stock ordering, etc. For example, if you were interested in using the rice sequence to identify candidate genes for a maize mutation, you could search for mutants in rice with a similar phenotype (using PO anatomy terms), then use Gramene's comparative map tool (CMAP) to locate the syntenic region in rice and maize.
Generating Pie Charts
NOTE: At this time, the Pie Chart function does not work in Safari.
If a PO term is fully expanded (i.e. all its children PO terms are displayed), a pie chart icon will appear at the end of the line (Figure 11A).
Figure 11A 
Clicking on the pie chart icon will generate a pie chart and a table showing the distribution of annotations associated with that PO term and its children (Figure 11).
Figure 11 
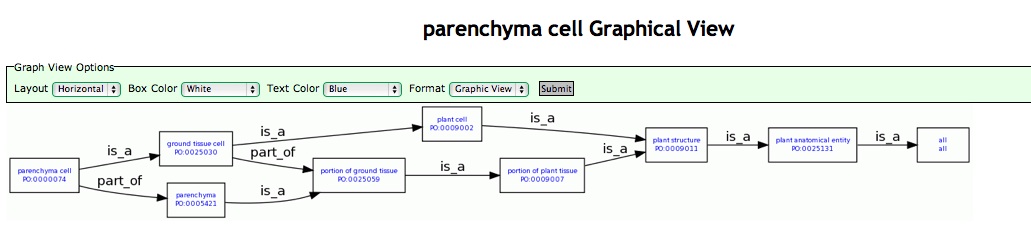
Graphical Ontology Views
A graphical view of each term and its lineage is linked from the term detail page. The header displays the name of the selected term. The remainder of the page displays a box and stick diagram of the term lineage. Each term in the lineage is contained in a box and terms are connected to each other by lines representing ontology relations. The arrowheads indicate the direction of the graph and are labeled with the relationship type between the term and its parent. The control panel below the term name can be used to change the layout of the graphical view and the color of the text and term boxes.
Figure 12