Introduction
AmiGO is an HTML based application which allows users to browse,query and visualize data from any ontology in OBO format. The AmiGO browser developed for GO has been modified for use on the Plant Ontology site. AmiGO is an open source software available from the go-dev cvs repository at the Gene Ontology SourceForge site.
Browser Compatibility
AmiGO is compatible with most browser/operating systems that support Cascading Style Sheets (CSS) including:
- FireFox/Mozilla/Netscape 6 and up on Linux, Windows and Mac OS X.
- Internet Explorer on windows but NOT Mac OS X. IE on Mac OS X does not support CSS standards.
- Safari on Mac OS X.
- Konquerer on Linux.
- Opera on Linux, Windows and Mac OS X.
The Main Window
From the AmiGO browser main page you have options to do the following:
- Browse the ontologies.
- Query the ontologies and/or associated gene products.
- Filter the gene products which are displayed in the browser.
- Perform an advanced search.
- Get the data in the current page in another format.
- Get data from a related website/database.
Browsing the ontologies.
Figure 1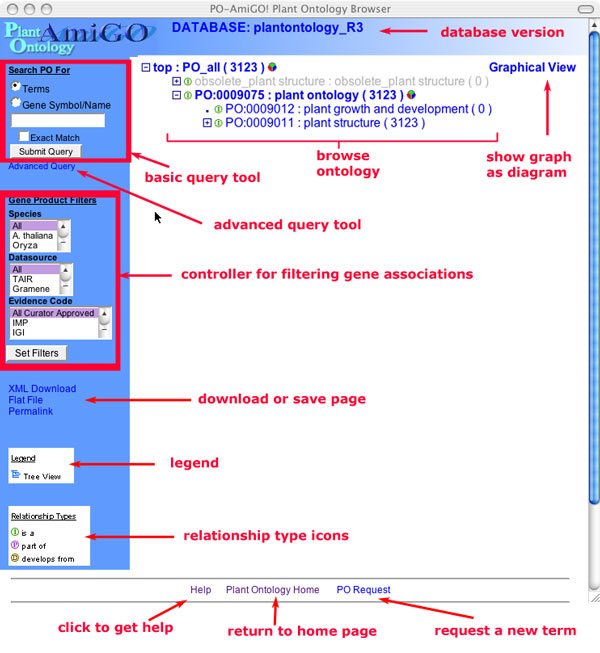
Database versioning
Each database release of the ontology and associated data is indicated by the version number in the upper right hand corner of the browser. The ontologies and associations are updated on a regular basis, and the data in the AmiGO browser will change accordingly.Therefore, different database versions will have different content. When performing queries, note the version number.
Ontology Tree Structure
The structure of the ontology is presented as a tree of terms. The top term or node is the highest level of the ontology.Each term is identified by its ontology ID which has a PO (Plant Ontology) prefix that indicates the ontology domain and a seven digit number (e.g. PO:0009075). This identifier is unique for each term in the ontology.
The level of a term in the ontology is represented by its indentation within the tree. In the Plant Ontology, plant structure (PO:0009011) is at the same level as plant growth and development (PO:0009012; data not yet shown), these terms are siblings. Furthermore, the relationship of a term to its parent is represented by the icon immediately preceding the term ID. Relationship types are: part of, represented by the pink P icon; isa, represented by a green I icon and develops from, represented by a brown D icon. In the plant ontology, both plant structure and plant growth and development have a 'part of' relationship to the parent term (plant ontology, PO:0009075).
Understanding term lines
Figure 2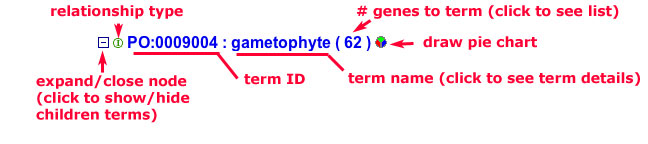
Each line in the tree contains one term and several pieces of information about the term and some associated functions.
Expanding/Closing term nodes: The term select/deselect icon has three forms: a plus sign (+), a minus sign (-) and a period (.). The plus sign indicates that a term has children that are not shown in the current view. The minus sign indicates the term is selected and all of the children are shown in the view. The period indicates that the selected term has no children. You can use the select/deselect icon to expand a node to see all the children, or collapse the node to just the parent term.
Term ID and Term name: The next two items are the term id and the term name which are separated by a semicolon. To see the definition of a term , move the cursor over the term and rest it for a moment or two. A small yellow window should pop-up containing the definition of the term. Depending on your browser, the definition may be truncated. To see the full definition of a term, click on the term name to see the detailed term view.
The status of a term is indicated by color and font style.If a term is selected in the tree, the name will be shown in bold font. Obsolete terms, shown in grey font, are retained and displayed in the browser because they are sometimes referred to by other terms or may have associations that have not been updated. In some cases an obsolete term ID is used as a secondary identifier for a newer term that was based on the older term.In the example shown in Figure 2a, the term "gametophyte" is selected (bold) and the term "obsolete_plant_structure" is shown in grey.
Number of annotations:The next item is a number in parenthesis. This number represents the total number of gene products or other types of data annotated to that term. This number can change depending on the search filters applied. When using some filters, the number of annotations cannot be accurately computed, therefore the number appears with a less than (<) sign. Point the cursor over the number in parenthesis to see an explanation of where the uncertainty comes from.
Browsing the tree
The select/deselect icon is used for browsing through the tree structure. If a term has children, you can 'drill down' by clicking on the plus sign selector next to the term. Terms with no children have a period (.). To deselect a node, click on the minus sign (-) to collapse the node and hide the children.
Querying the database and/or associated gene products.
The ontology can be queried using the search options available in the upper left hand side of the screen.
Figure 3
The database query works by entering a query string into the text input box and selecting the type of search to perform (exact/inexact) and data type to search for. The default option is a search for terms, meaning a search for the term 'leaf' will retrieve all terms containing the word 'leaf'.
Querying for terms
The text string used for queries treats each word in the string as a complete phrase. A search for the term "juvenile leaf" will find terms containing the string "juvenile leaf" rather than "juvenile" OR "leaf". For the broadest search use a simple term and the contains option. In addition to searching the term names, the term search also searches alternative names, such as synonyms, which are stored in the database. For example, a search for the term 'panicle' which is the name of the inflorescence in rice, will return the term 'inflorescence' because panicle is a synonym for this term.
- Enter the term you are searching for into the text input box.
- If you know the exact term you are searching for, choose the "Exact Match" option.
- Select terms as the data type to search for. This is the default selected option.
- Click 'Submit Query'.
The results that match your query are shown in a list. If no results are found, check your spelling or try using simpler terms (leaf vs. juvenile leaf). If you do not find a term in the PO, you can suggest a new term by clicking on the "PO Request" link at the bottom of the browser window.
Figure 4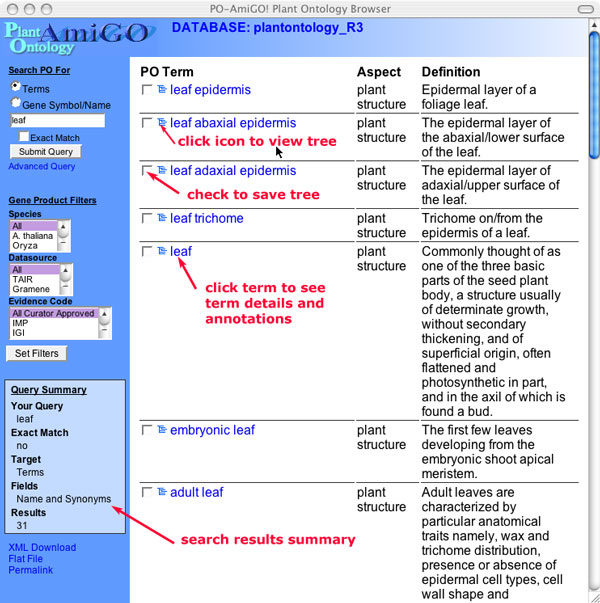
Click on the name of the term to display the term detail view.To the left of the term are a select box and an icon representing an ontology tree. To view the tree structure showing the terms location within the ontology (its parentage all the way to the top most node),click on the tree icon. If more than one term is shown in the results, you can check the terms you want to display in the tree view and choose the "Show Checked items in tree" option. Multiple tree views can be shown. Additional views can be added by selecting the term and choosing the "Add checked items to current tree" option.
Querying for gene products
AmiGO allows you to search for gene products or other data types associated to terms in the database. As with the term search, names and aliases (alternative names for the gene) are included in the search.
Figure 5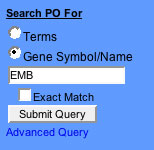
- Enter the name (symbolic or full name ) of the gene product such as EMB.
- If you know the exact name, choose an 'Exact Match' search, other wise the search will be a 'contains' search.
- Choose the "Gene Symbol" option.
- Click "Submit Query".
The query for gene names containing the word "EMB" is shown in Figure 6.
Figure 6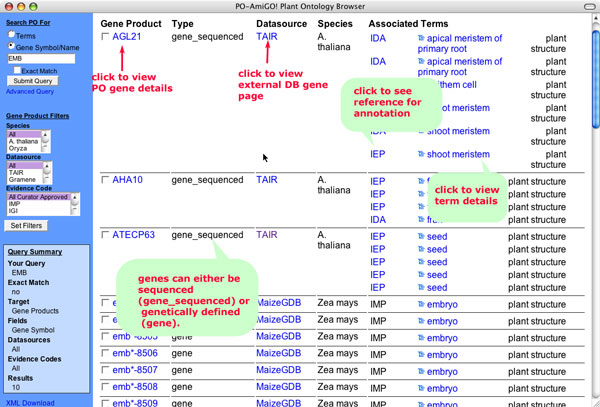
The Query Summary, shown in the lower left hand box lists the search parameters,data filtering options selected and number of results returned. The main results section lists all of the gene products that matched your query. To the left of each result is a check box which can be used for selecting items on the list. Selected results can be shown in a detailed viewor as a set of FASTA files (not yet available).
Gene Product name: This field displays the primary name for the gene product as it is stored in the database. Remember that the gene search includes aliases, so the name which appears in the results may not match the query term. Click on the term name to view the gene detail view.
Type: There are currently two types of genes in the Plant Ontology database. gene_sequenced refers to genes which have been sequenced. The type 'gene' refers to genetically defined loci which may or may not be mapped to a linkage group. Genetically defined loci include characterized mutations that segregate as a single locus as well as quantitative trait loci (QTL).
Datasource: This refers to the database which is the source of the annotation. Datasources include Gramene (rice and other grasses, TAIR (Arabidopsis), and MaizeGDB (maize). This is one of the filtering options available.
Species: The species in which the gene product is found, is indicated in this field.Each database may contribute annotations to more than one species. The species option is also available as a filtering option to select only species of interest in the results.
Associated Terms: For each gene product, the terms to which it is annotated to, and the supporting evidence code are listed, one gene may have many associated terms. To see what each evidence code means, point the cursor over the three-letter code. A yellow box will pop-up showing the complete evidence term (e.g., IMP=Inferred from mutant phenotype). See PO evidence code documentation for a complete explanation of the codes and their usage. To view the term details, click on the term name. Click on the tree icon next to the term name to see the tree view centered on the term. The evidence codes are hyperlinked to the publication or other resource that is the source of evidence supporting the assertion. Annotations may appear to be redundant if more than one evidence code or source of evidence is used to make the association of a gene to a term.
Aspect: The last column in the list of results shows the aspect of the ontology to which the term belongs. In the plant ontology, this will either be plant structure or plant growth and development.
Filtering gene products which are displayed in the browser
AmiGO has three filters which you can use to filter the list of gene products displayed in the browser or included in search results. The default option does not use any filters for retrieving associated data. The filters can be set when performing an advanced database search, or when viewing the ontology tree or term details.One or more filters can be set; the corresponding results will meet the criteria set by ALL the filters in combination.
Using the species filter
The species filter allows you to restrict the display of annotated data in the browser display or search results to one or more species. This feature is useful if you want to find annotations to a specific organism (say Zea mays) or a set of organisms. For example, say you wanted to take advantage of maize-rice synteny for identifying candidate genes in maize. Given a phenotype in maize, are there mutants in rice with similar phenotypes?
Figure 7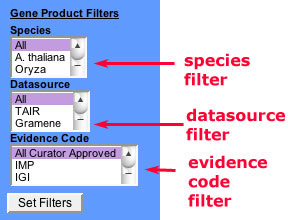
- Select one or more species to filter. To select multiple species, hold down the CTRL key (PC) or APPLE key (Mac) while selecting with the mouse.
- Click 'Set Filters'.
Ontology view: When the filters are set on the ontology (tree view), the number of gene products (or other data types) associated with each term will change. If the exact number of genes cannot be determined, the value will be shown with a < , indicating that fewer than the total number of genes from the database will be included in the list of genes obtained by clicking on this link.
Term Detail view: When filters are applied to term detail views, the list of genes (or other annotated data) included on the term detail view will be limited to only the selected species.
Using the datasource filter
This portion allows you to restrict the display of annotated data by contributing database. It functions similarly to the species filter by limiting the associated data types in the ontology and term detail views as well as in searches.
- Select the datasources to include by clicking with the mouse. To choose multiple datasources, hold down the CTRL key (PC) or APPLE key (Mac) while making your selections.
- Click 'Set Filters'.
Ontology view: Setting filters when viewing the ontology will change the number of associated data objects to reflect the data contributed by the selected datasources.
Term Detail view: Setting the database filter will redraw the page to contain only the associated data from the selected datasource.
Using the evidence code filter
The evidence code filter is a very useful option that allows you to restrict the display of associated data based on the quality of evidence supporting the annotation. The default option is set to 'All Curator Approved'. Curator approved means the association of a term to a data object is based on evidence that was evaluated by a trained biologist. This default option excludes annotations based on electronic unsupervised methods.
Performing an advanced search
The advanced search features are accessible by clicking on the 'Advanced Search' link below the main query box in the AmiGO toolbar. These options allow you to query the PO in bulk and to specify the exact type of terms to search. It can also be used to filter the gene associations that are returned.
Figure 8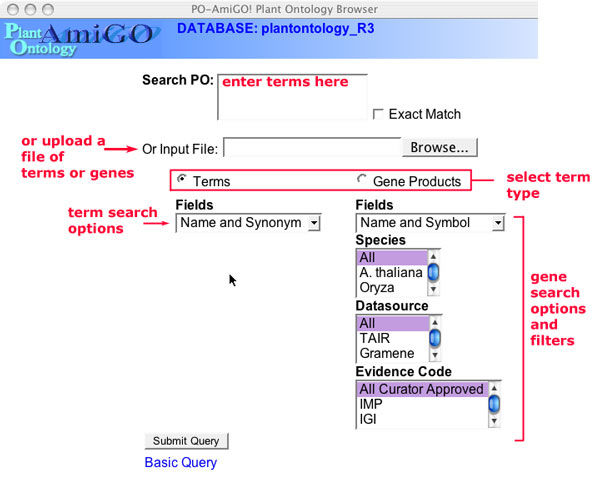
Entering query terms. In the text input box you can enter a single word as with the Basic Query; you can also enter multiple terms separated by new lines (carriage returns) and AmiGO will search for hits to each of these terms. Alternatively, you can upload a file of terms where each term is on a single line. The terms must all be of the same type. Choose 'Terms' or 'Gene Products' to search one or the other of these fields.
- Enter the term(s) in the text input box or upload a file of query terms.
- Choose the term type to search, the choices are terms or Gene products. Choose an exact or inexact match. For exact matches, check the "Exact Match" box.
- If searching for gene products, set the filtering options.
- Click "Submit Query" .
When searching for terms, the types of terms you can search for are:
- Term Name and Synonym.
- Term Name and Definition.
- Ontology ID: e.g. PO:0009074
- All of the above fields
When searching for gene products, the options are:
- Gene Full Name and Symbolic Name
- Full Gene Name
- Sequence Accession (only for sequenced genes)
- Gene Symbol
- Synonyms (Gene Name aliases)
Getting the data in the current page in another format
XML or Flat File: Clicking on either of these links will bring you to a new page with a different representation of a selected ontology tree. This option is useful if you want to parse the tree using either the XML or GO flat file format. Some browsers (such as Safari) do not display XML properly.
Permalinks: Clicking on this option creates a new page with a new URL which you can use to bookmark the currently selected tree. AmiGO tracks the tree you build in your 'user session'. After 2 hours if inactivity, the session will expire along with any trees you have built. To save the tree indefinitely, add the URL from the Permalink to your browsers bookmarks file.
Getting data from a related website/database.
There are several places where you can find links to related databases or websites to find additional information about a gene or term.
Datasource links
The term detail pages and gene detail pages include hyperlinks to data detail pages from the contributing database. The datasource listed in this field (e.g. TAIR, Gramene or MaizeGDB) is a hyperlink. Click on this link to view the databases detail page for the gene, phenotype, allele or other data type. For each database, the detail pages will provide as much information as they have available about the data type such as sequences, map locations, stock ordering, etc... For example, if you were interested in using the rice sequence to identify candidate genes for a maize mutation you could search for mutants in rice with a similar phenotype (using PO anatomy terms), then use Gramene's comparative map tool (C-MAP) to locate the syntenic region in rice and maize.
Evidence Links
The evidence codes on the annotation display on the term detail pages may be hyperlinks. These hyperlinks go to the corresponding record in the database, which supplied the annotation, where you can find information about the experimental method which was used to support the annotation. For example, a gene may be annotated to an anatomy term (e.g., microgametophyte) because a mutation in the gene is male sterile. The evidence code would then be "IMP" or inferred from mutant phenotype and the hyperlink may go to a reference describing the mutant phenotype. You can use these links to find out more information about the gene product and to verify the annotation yourself.
Detail Windows
There are detail pages for terms and gene products that are in the PO database. These pages are displayed in smaller browser windows when you click on the name of a term or a gene product. These pages open in new windows, which also allows you to view a tree that you have been browsing in the main window open, while exploring interesting terms and gene products in depth.
Term Details
The term detail page is linked from the term names in the main tree view, from the term name in the search results page, and the term names in the gene product detail pages.
Figure 9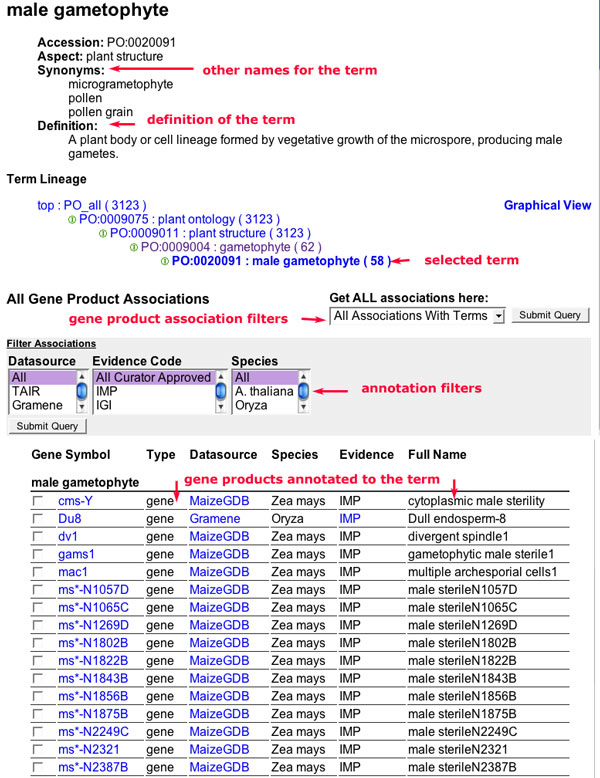
The top of the page shows the name of the term. Following that, are the term accession, synonyms and definition.The accession is a unique identifier for the term in the PO database. Synonyms are alternative names for the term. In PO, synonyms may include more specific instances of a term. For example, a cyme is a specific type of inflorescence and is included as a synonym for the term inflorescence.
Term Lineage
The next section on the detail page is the Term Lineage. This section shows the tree view (actually a directed acyclic graph or DAG) of the term. This view is similar to the main tree view but without the select/deselect icons. Next to the term lineage header is a link to a graphical view of the term.
Gene Product Associations
The rest of the page is taken up by the gene product associations and filters for viewing subsets of the associations.The selector to the right of this header can be used to determine the associated genes to display.
- Select the associations to display. The options are all associations, direct associations and all associations with terms.
- Click "Submit Query".
The header of this section will say one of three things.
All Gene Product Associations with terms:
By default, all of the gene products associated to the selected term AND all of its children are shown in the term detail view. The display shows all of the descendant terms and the genes associated to each of the terms. The number of gene products displayed in this view should be equal to the number shown in parenthesis following the term name in the tree view.
Direct Gene Product Associations:
These are gene products which have been associated DIRECTLY to the selected term.
All Associations:
This option displays all of the associations to the term and its descendants without specifying which terms the gene products are annotated to. This option is useful if you want to download sets of genes that fall within a broad category (e.g., all genes expressed in the root regardless of tissue or cell type.)
The following is a description of each of the fields displayed in the Gene Association section. If the terms are displayed, the genes associated to each of the terms is shown in the corresponding section.
- Gene Symbol: This is the symbolic name for the gene associated to the term. This is hyperlinked to the gene detail page.
- Type: This is the type of gene product. Options are gene or sequenced gene.
- Datasource: This refers to the source of the information about the gene and the annotation. This is hyperlinked to the corresponding data detail pages from the contributing database.
- Species: The species from which the gene product is derived.
- Evidence: The three letter code corresponding to the evidence supporting the annotation. The evidence code may be linked to the reference in the contributing database.
- Full name: The full name of the gene product is shown in this field. Full name may be the complete name of a mutant (e.g. barren sterile 1) or the gene product itself (e.g. catalase 1).
Filtering options
The gene association filters can be used to select a subset of gene associations to display. Select one or more filters to apply and the click on the "submit query" button. The term detail page will be redrawn and only include the selected gene associations. The default view does not restrict the associations.
Gene Details
The Gene Product Detail pages are linked from the gene product query results page and from the term detail pages. A typical gene page has the following information.
Figure 10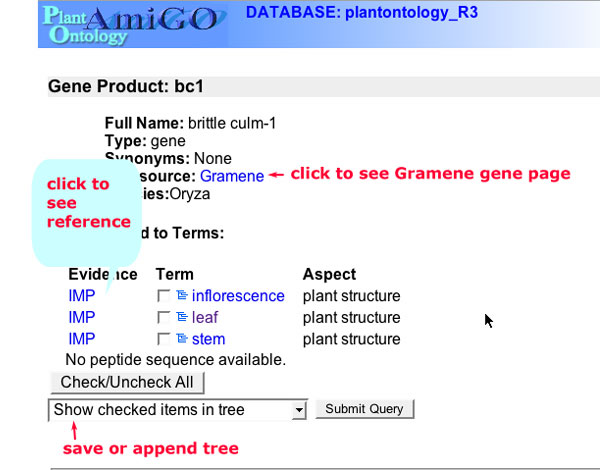
Gene product names, synonyms and datasources.
Each contributing database (datasource) submits information about the symbolic name, full name and aliases for each gene product. This information is displayed in this section and hyperlinked to the corresponding detail page in the contributing database.
Associated to Terms
Any gene product can be associated to one or more terms in the PO database. The term associations are shown in this section. The first item in the list is the evidence code which can be used to assay the strength of the assertion. The next item is a checkbox next to the term. The check box allows you to select one or more terms to add to a tree or append to an existing tree of terms. The tree can be saved as a permanent link and used to view or download sets of associations. The last item is the term the gene product is annotated to.The term name is hyperlinked to the term detail page.
Peptide Sequence
If the sequence of the gene product has been submitted to the PO database, the amino acid sequence will be displayed in this section.
Using the links out to contributing databases
Generating Pie Charts
Pie charts that summarize the distribution of gene products annotated to terms can be obtained by clicking on the pie chart icon in the tree view. The pie chart will display a summary of gene products annotated to the selected term and ALL OF ITS DESCENDANTS WHICH HAVE BEEN SELECTED. The number of selected products can be limited using the filters .The top of the pie chart shows the name of the selected term. Below the pie chart is a list summarizing the results. The first column displays the names of the selected term and all of the selected dependent terms. The next row is labeled "ALL" and lists the total number of gene products associated to the term and its descendants (selected or otherwise). The second column shows the number of gene products associated to the term listed in the first column. The last column shows this number as a percentage of the number in the first row. This percentage represents the number of genes annotated to the term in the first column over the number of genes annotated to the selected (parent) term and all of its descendants (selected or not).
Figure 11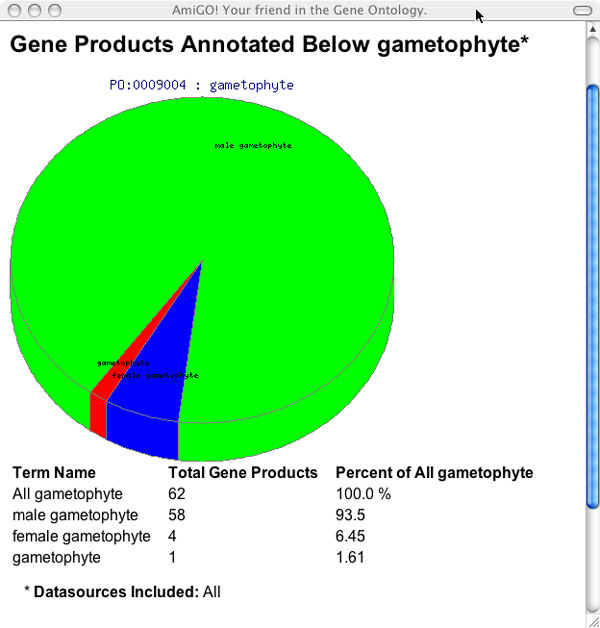
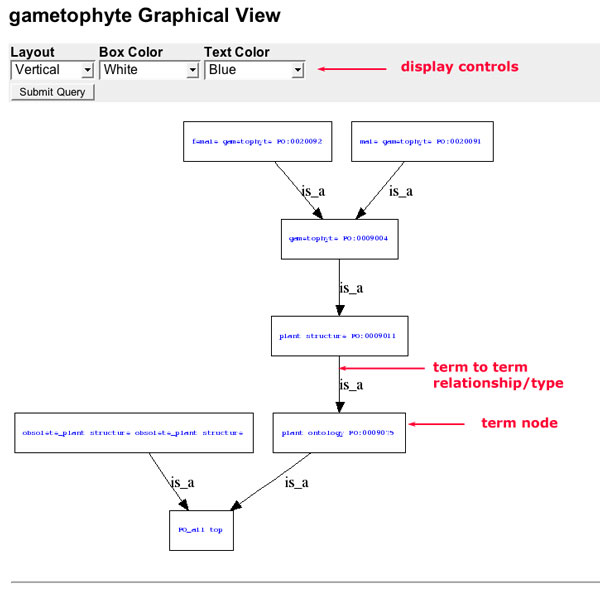
Graphical Ontology Views
A graphical view of each term and its lineage is linked from the term detail page. The header displays the name of the selected term. The remainder of the page displays a box and stick diagram of the term lineage where each term in the lineage is contained in a box and terms are connected to each other by lines. The arrowheads indicate the direction of the graph and are labeled with the relationship type between the term and its parent. The control panel below the term name can be used to change the layout of the graphical view and color of the text and term boxes.
Figure 12